SR24.txt
The taxpayers through the United State Department of Agriculture Agriculture Research Service, Nutrient Data Laboratory have provided a USDA National Nutrient Database for Standard Reference, Release 24 of foods, food ingredients, and nutrition. The current revision is 24.
These are notes on creating this database in Ubuntu 10.4 LTS Linux using SQLite 3.6.22.
These notes apply specifically to SR24.
I had prepared detailed notes using SR23 and tried to test the transition and discovered some differences that had I read the read the fine documentation…
NUT_DATA has a “AddMod_Date” added as a 17th column pushing CC to 18th.
Line 37 of DERIV_CD contains new-lines and threw sed off because I did not handle the new-lines within the quotes. As a temporary fix, hand edit the unzipped DERIV_CD.txt file to combine the long text into a single line before importing the data for DERIV_CD. Delete a new-line at the end of line 38 and end of line 37.
Google for: sr24 nutrition database
or navigate to:
http://www.ars.usda.gov/Services/
Select “datasets” from the menu at right
“Food and Nutrition” from the listed dataset categories
“Nutrient Data Laboratory” under Food and Nutrition
“USDA National Nutrient Database for Standard Reference, Release 24”
Download the ASCII version of the file.
Also download any patch versions if required.
Unzip the downloaded file.
unzip sr24.zip
If you had separately downloaded sr24_doc.pdf, the zip file version is identical.
Look at the PDF.
Table 3 on page 24 shows the names of the tables. There is a .txt file that corresponds to each of the tables.
Page 25 describes the tables and their relations. Figure 1 shows the relationships graphically.
Next come Tables 4 through 15 describing the data columns in each of the tables.
Look at one of the tables. I chose FOOTNOTE (Table 13) because it has a relatively small number of columns.
head FOOTNOTE.txt
~02009~^~01~^~D~^~~^~Mix of chili pepper, other spices and salt~
~02030~^~01~^~N~^~204~^~Total proximates do not equal 100% because piperine was subtracted from lipid value.~
~02032~^~01~^~N~^~204~^~Total proximates do not equal 100% because piperine was subtracted from lipid value.~
~02033~^~01~^~D~^~~^~Other phytosterols = 38.4 mg/100g; these include delta 5-avenasterol (17.7), campestanol (2.6), and other minor phytosterols (18.1 mg).~
~02034~^~01~^~N~^~204~^~Total proximates do not equal 100% because piperine was subtracted from lipid value.~
~02048~^~01~^~D~^~~^~Contains 5% acetic acid. Total proximates do not equal 100%.~
~02048~^~02~^~N~^~208~^~Acetic acid is included in energy calculation.~
~02053~^~01~^~D~^~~^~Contains 5% acetic acid. Total proximates do not equal 100%.~
~02053~^~02~^~N~^~208~^~Acetic acid is included in energy calculation.~
~02063~^~01~^~D~^~~^~Leaf~
Each field is “quoted” with tildes and separated with a carat. Fields with null values have adjacent tildes (02048 and 02063 are examples).
Compare the text with the description in Table 13.
Table 13.—Footnote File Format
|
Field |
Type |
Size |
Blank |
Description |
|
|
NDB_No |
A |
5 |
N |
5-digit |
|
|
Footnt_No |
A |
4 |
N |
Sequence |
|
|
Footnt_Typ |
A |
1 |
N |
Type |
|
|
D |
|||||
|
M |
|||||
|
N |
|||||
|
Nutr_No |
A |
3 |
Y |
Unique |
|
|
Footnt_Txt |
A |
200 |
N |
Footnote |
|
For example NDB_No is a 5 digit numeric (as characters) field including leading zeros.
Footnt_Txt is a 200 character field
Nutr_no, if not blank is a 3-digit identifier of the nutrient to which the footnote applies.
You can similarly do each of the other tables.
There are 3 steps required for each table:
1. Edit the .txt file to remove the tildes.
2. Create the tables in an SQLlite database
3. Import the data from the edited .txt files
The next step is create, define and load the database. SQLite3 is available on the Ubuntu system but may not be initially loaded.
This can be tested by typing sqlite3 at a command prompt. If the program starts, use .quit to end it (;.quit if you are in the middle of SQL). .help will provide a command summary of the .commands. Ubuntu will tell you to use “sudo apt-get install sqlite3” if the program is not yet present. Using apt-get will install the program if necessary. Other Linux system distributions may offer similar help but may have different installation tools. SQLite3 is controlled by a combination of .commands and SQL.
Edit the text file for DERIV_CD to combine that long line.
Line 37 of DERIV_CD contains new-lines. Hand edit the unzipped DERIV_CD.txt file to combine the long text into a single line before importing the data for DERIV_CD. Delete a new-line at the end of line 38 and end of line 37.
You should be able to copy-and-paste the text from “here” to “there” (excluding here and there) and paste it into your Linux command prompt.
here mkdir temp sed -e 's/~//g' DATA_SRC.txt > temp/DATA_SRC.txt sed -e 's/~//g' DERIV_CD.txt > temp/DERIV_CD.txt sed -e 's/~//g' FOOD_DES.txt > temp/FOOD_DES.txt sed -e 's/~//g' LANGDESC.txt > temp/LANGDESC.txt sed -e 's/~//g' NUT_DATA.txt > temp/NUT_DATA.txt sed -e 's/~//g' SRC_CD.txt > temp/SRC_CD.txt sed -e 's/~//g' DATSRCLN.txt > temp/DATSRCLN.txt sed -e 's/~//g' FD_GROUP.txt > temp/FD_GROUP.txt sed -e 's/~//g' FOOTNOTE.txt > temp/FOOTNOTE.txt sed -e 's/~//g' LANGUAL.txt > temp/LANGUAL.txt sed -e 's/~//g' NUTR_DEF.txt > temp/NUTR_DEF.txt sed -e 's/~//g' WEIGHT.txt > temp/WEIGHT.txt ls -l temp rm SR24.db sqlite3 SR24.db .tables CREATE TABLE ABBREV(NDB_No INTEGER, Shrt_Desc, Water FLOAT, Energ_Kcal FLOAT, Protein FLOAT, Lipid_Tot FLOAT, Ash FLOAT, Carbohydrt FLOAT, Fiber_TD FLOAT, Sugar_Tot FLOAT, Calcium FLOAT, Iron FLOAT, Magnesium FLOAT, Phosphorus FLOAT, Potassium FLOAT, Sodium FLOAT, Zinc FLOAT, Copper FLOAT, Manganese FLOAT, Selenium FLOAT, Vit_C FLOAT, Thiamin FLOAT, Riboflavin FLOAT, Niacin FLOAT, Panto_acid FLOAT, Vit_B6 FLOAT, Folate_Tot FLOAT, Folic_Acid FLOAT, Food_Folate FLOAT, Folate_DFE FLOAT, Vit_B12 FLOAT, Vit_A_IU FLOAT, Vit_A_RAE FLOAT, Retinol FLOAT, Vit_E FLOAT, Vit_K FLOAT, Alpha_Carot FLOAT, Beta_Carot FLOAT, Lycopene FLOAT, Lut_Zea FLOAT, FA_Sat FLOAT, FA_Mono FLOAT, FA_Poly FLOAT, Cholestrl FLOAT, GmWt_1 FLOAT, GmWt_Desc1, GmWt_2 FLOAT, GmWt_Desc2, Frefuse_Pct INTEGER); CREATE TABLE DATA_SRC(DataSrc_ID INTEGER, Authors, Title, Year INTEGER, Journal, Vol_City, Issue_State, Start_Page, End_Page); CREATE TABLE DATSRCLN(NDB_No INTEGER, Nutr_No INTEGER, DataSrc_ID INTEGER); CREATE TABLE DERIV_CD(Deriv_Cd INTEGER, Deriv_Desc); CREATE TABLE FD_GROUP(FdGrp_Cd INTEGER, FdGrp_Desc); CREATE TABLE FOOD_DES(NDB_No INTEGER PRIMARYKEY, FdGrp_Cd INTEGER, Long_Desc VARCHAR(200), Shrt_Desc VARCHAR(60), ComName VARCHAR(100), ManufacName VARCHAR(50), Survey CHAR(1), Ref_desc VARCHAR(60), Refuse INTEGER, SciName VARCHAR(60), N_Factor FLOAT, ProFactor FLOAT, FatFactor FLOAT, CHO_Factor FLOAT); CREATE TABLE FOOTNOTE(NDB_No INTEGER, Footnt_No INTEGER, Footnt_Typ, Nutr_No INTEGER, Footnt_Txt); CREATE TABLE NUTR_DEF(Nutr_No INTEGER, Units, Tagname, NutrDesc, Num_Dec INTEGER, SR_Order INTEGER); CREATE TABLE NUT_DATA(NDB_No INTEGER, Nutr_No INTEGER, Nutr_Val FLOAT, Num_Data_Pts FLOAT, Std_Error FLOAT, Src_Cd, Deriv_Cd, Ref_NDB_No INTEGER, Add_Nutr_Mark, Num_Studies INTEGER, Min FLOAT, Max FLOAT, DF INTEGER, Low_EB FLOAT, Up_EB FLOAT, Stat_cmt, AddMod_Date, CC); CREATE TABLE SRC_CD(Src_Cd INTEGER, SrcCd_Desc); CREATE TABLE WEIGHT(NDB_No INTEGER, Seq INTEGER, Amount FLOAT, Msre_Desc, Gm_Wgt FLOAT, Num_DataPts INTEGER, Std_Dev FLOAT); CREATE TABLE LANGUAL(NDB_No INTEGER, Factor_Code); .tables .separator ^ .import temp/DATSRCLN.txt DATSRCLN .import temp/FOOD_DES.txt FOOD_DES .import temp/LANGUAL.txt LANGUAL .import temp/FOOTNOTE.txt FOOTNOTE .import temp/NUT_DATA.txt NUT_DATA .import temp/WEIGHT.txt WEIGHT .import temp/DATA_SRC.txt DATA_SRC .import temp/DERIV_CD.txt DERIV_CD .import temp/FOOD_DES.txt FOOD_DES .import temp/NUTR_DEF.txt NUTR_DEF .import temp/SRC_CD.txt SRC_CD .quit there
I should have mentioned that some of those files are large with perhaps a long pause between steps.
ABBREV is a cut-down version of the entire database and is made to be used stand-alone.
Relationships
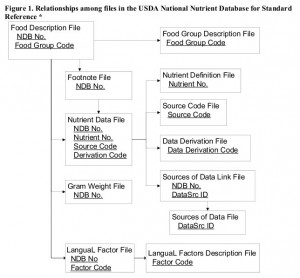
Table Schema
ABBREV
*NDB_No INTEGER
Shrt_Desc
Water FLOAT
Energ_Kcal FLOAT
Protein FLOAT
Lipid_Tot FLOAT
Ash FLOAT
Carbohydrt FLOAT
Fiber_TD FLOAT
Sugar_Tot FLOAT
Calcium FLOAT
Iron FLOAT
Magnesium FLOAT
Phosphorus FLOAT
Potassium FLOAT
Sodium FLOAT
Zinc FLOAT
Copper FLOAT
Manganese FLOAT
Selenium FLOAT
Vit_C FLOAT
Thiamin FLOAT
Riboflavin FLOAT
Niacin FLOAT
Panto_acid FLOAT
Vit_B6 FLOAT
Folate_Tot FLOAT
Folic_Acid FLOAT
Food_Folate FLOAT
Folate_DFE FLOAT
Vit_B12 FLOAT
Vit_A_IU FLOAT
Vit_A_RAE FLOAT
Retinol FLOAT
Vit_E FLOAT
Vit_K FLOAT
Alpha_Carot FLOAT
Beta_Carot FLOAT
Lycopene FLOAT
Lut_Zea FLOAT
FA_Sat FLOAT
FA_Mono FLOAT
FA_Poly FLOAT
Cholestrl FLOAT
GmWt_1 FLOAT
GmWt_Desc1
GmWt_2 FLOAT
GmWt_Desc2
Frefuse_Pct INTEGER
DATA_SRC
*DataSrc_ID INTEGER
Authors
Title
Year INTEGER
Journal
Vol_City
Issue_State
Start_Page
End_Page
DATSRCLN
*NDB_No INTEGER
Nutr_No INTEGER
DataSrc_ID INTEGER
DERIV_CD
*Deriv_Cd INTEGER
Deriv_Desc
FD_GROUP
FdGrp_Cd INTEGER
FdGrp_Desc
FOOD_DES
*NDB_No INTEGER PRIMARYKEY
FdGrp_Cd INTEGER
Long_Desc VARCHAR(200)
Shrt_Desc VARCHAR(60)
ComName VARCHAR(100)
ManufacName VARCHAR(50)
Survey CHAR(1)
Ref_desc VARCHAR(60)
Refuse INTEGER
SciName VARCHAR(60)
N_Factor FLOAT
ProFactor FLOAT
FatFactor FLOAT
CHO_Factor FLOAT
FOOTNOTE
*NDB_No INTEGER
Footnt_No INTEGER
Footnt_Typ
Nutr_No INTEGER
Footnt_Txt
NUTR_DEF
*Nutr_No INTEGER
Units
Tagname
NutrDesc
Num_Dec INTEGER
SR_Order INTEGER
NUT_DATA
*NDB_No INTEGER
Nutr_No INTEGER
Nutr_Val FLOAT
Num_Data_Pts FLOAT
Std_Error FLOAT
Src_Cd
Deriv_Cd
Ref_NDB_No INTEGER
Add_Nutr_Mark
Num_Studies INTEGER
Min FLOAT
Max FLOAT
DF INTEGER
Low_EB FLOAT
Up_EB FLOAT
Stat_cmt
AddMod_Date
CC
SRC_CD
*Src_Cd INTEGER
SrcCd_Desc
WEIGHT
*NDB_No INTEGER
Seq INTEGER
Amount FLOAT
Msre_Desc
Gm_Wgt FLOAT
Num_DataPts INTEGER
Std_Dev FLOAT
Sample Queries
Some sample queries and results – copy and paste the query to the sqlite> prompt.
SELECT * FROM FD_GROUP;
100|Dairy and Egg Products
200|Spices and Herbs
300|Baby Foods
400|Fats and Oils
500|Poultry Products
600|Soups, Sauces, and Gravies
700|Sausages and Luncheon Meats
800|Breakfast Cereals
900|Fruits and Fruit Juices
1000|Pork Products
1100|Vegetables and Vegetable Products
1200|Nut and Seed Products
1300|Beef Products
1400|Beverages
1500|Finfish and Shellfish Products
1600|Legumes and Legume Products
1700|Lamb, Veal, and Game Products
1800|Baked Products
1900|Sweets
2000|Cereal Grains and Pasta
2100|Fast Foods
2200|Meals, Entrees, and Sidedishes
2500|Snacks
3500|Ethnic Foods
Specify a specific row using SQL expressions
SELECT * FROM FD_GROUP WHERE FdGrp_Cd=600
...> ;
600|Soups, Sauces, and Gravies
Do not forget the semi-colon terminator. If you do the …> prompt will return until you do.
When you select *, you get all of the values in each row. They will be separated by the default separator | or the most recently specified .separator value.
SELECT * FROM FOOD_DES WHERE FdGrp_Cd=3500;
35001|3500|Agutuk, fish/berry with seal oil (Alaskan ice cream) (Alaska Native)|AGUTUK,FISH/BERRY W/ SEAL OIL (ICE CREAM) (ALASKA NATIVE)|||||0||6.25|||
35002|3500|Agutuk, fish with shortening (Alaskan ice cream) (Alaska Native)|AGUTUK,FISH W/ SHORTENING (ALASKA NATIVE)|||||0||6.25|||
35003|3500|Agutuk, meat-caribou (Alaskan ice cream) (Alaska Native)|AGUTUK,MEAT-CARIBOU (ALASKAN ICE CREAM) (ALASKA NATIVE)|||||0||6.25|||
...
A more complex query specifying specific selection and columns to return
SELECT FOOD_DES.NDB_No, FOOD_DES.Shrt_Desc, NUT_DATA.Nutr_No, NUT_DATA.Nutr_Val, NUTR_DEF.Units, NUTR_DEF.NutrDesc FROM FOOD_DES, NUT_DATA, NUTR_DEF WHERE ((FOOD_DES.FdGrp_Cd=3500) AND ((FOOD_DES.NDB_No>35130) AND (FOOD_DES.NDB_No<35132)) AND ((NUTR_DEF.Nutr_No>300) AND (NUTR_DEF.Nutr_No<350)) AND (NUT_DATA.Nutr_No=NUTR_DEF.Nutr_No) AND (FOOD_DES.NDB_No=NUT_DATA.NDB_No));
35131|CORNMEAL,BLUE (NAVAJO)|301|5.0|mg|Calcium, Ca
35131|CORNMEAL,BLUE (NAVAJO)|303|2.91|mg|Iron, Fe
35131|CORNMEAL,BLUE (NAVAJO)|304|133.0|mg|Magnesium, Mg
35131|CORNMEAL,BLUE (NAVAJO)|305|354.0|mg|Phosphorus, P
35131|CORNMEAL,BLUE (NAVAJO)|306|393.0|mg|Potassium, K
35131|CORNMEAL,BLUE (NAVAJO)|307|7.0|mg|Sodium, Na
35131|CORNMEAL,BLUE (NAVAJO)|309|2.91|mg|Zinc, Zn
35131|CORNMEAL,BLUE (NAVAJO)|312|0.218|mg|Copper, Cu
35131|CORNMEAL,BLUE (NAVAJO)|315|0.758|mg|Manganese, Mn
35131|CORNMEAL,BLUE (NAVAJO)|317|11.8|μg|Selenium, Se
35131|CORNMEAL,BLUE (NAVAJO)|323|0.73|mg|Vitamin E (alpha-tocopherol)
35131|CORNMEAL,BLUE (NAVAJO)|341|0.0|mg|Tocopherol, beta
35131|CORNMEAL,BLUE (NAVAJO)|342|3.21|mg|Tocopherol, gamma
35131|CORNMEAL,BLUE (NAVAJO)|343|0.37|mg|Tocopherol, delta
35131|CORNMEAL,BLUE (NAVAJO)|301|5.0|mg|Calcium, Ca
35131|CORNMEAL,BLUE (NAVAJO)|303|2.91|mg|Iron, Fe
35131|CORNMEAL,BLUE (NAVAJO)|304|133.0|mg|Magnesium, Mg
35131|CORNMEAL,BLUE (NAVAJO)|305|354.0|mg|Phosphorus, P
35131|CORNMEAL,BLUE (NAVAJO)|306|393.0|mg|Potassium, K
35131|CORNMEAL,BLUE (NAVAJO)|307|7.0|mg|Sodium, Na
35131|CORNMEAL,BLUE (NAVAJO)|309|2.91|mg|Zinc, Zn
35131|CORNMEAL,BLUE (NAVAJO)|312|0.218|mg|Copper, Cu
35131|CORNMEAL,BLUE (NAVAJO)|315|0.758|mg|Manganese, Mn
35131|CORNMEAL,BLUE (NAVAJO)|317|11.8|μg|Selenium, Se
35131|CORNMEAL,BLUE (NAVAJO)|323|0.73|mg|Vitamin E (alpha-tocopherol)
35131|CORNMEAL,BLUE (NAVAJO)|341|0.0|mg|Tocopherol, beta
35131|CORNMEAL,BLUE (NAVAJO)|342|3.21|mg|Tocopherol, gamma
35131|CORNMEAL,BLUE (NAVAJO)|343|0.37|mg|Tocopherol, delta
In the output above, I have substituted a μ for the unknown character � that appears on the Linux console where the query was run. μ is not in the console font.
Curious as to why the result was duplicated?
SELECT FOOD_DES.Shrt_Desc FROM FOOD_DES WHERE FOOD_DES.NDB_No=35131;
CORNMEAL,BLUE (NAVAJO)
CORNMEAL,BLUE (NAVAJO)
The ingredient description is duplicated.
SELECT * FROM FOOD_DES WHERE FOOD_DES.NDB_No=35131;
35131|3500|Cornmeal, blue (Navajo)|CORNMEAL,BLUE (NAVAJO)|||||0|||||
35131|3500|Cornmeal, blue (Navajo)|CORNMEAL,BLUE (NAVAJO)|||||0|||||
It may be helpful to add the SQL keyword DISTINCT to queries
SELECT DISTINCT * FROM FOOD_DES WHERE FOOD_DES.NDB_No=35131;
35131|3500|Cornmeal, blue (Navajo)|CORNMEAL,BLUE (NAVAJO)|||||0|||||
